The overall objective is to study the effect of spatial gradients and Nostoc availability on interaction networks of the Peltigera–Nostoc lichen symbiosis. More specifically, we will address these goals:
Goal 1a. What is the spatial scale, and associated environmental factors, of Nostoc phylogroup replacement, and how does this relate to Peltigera–Nostoc network structure?
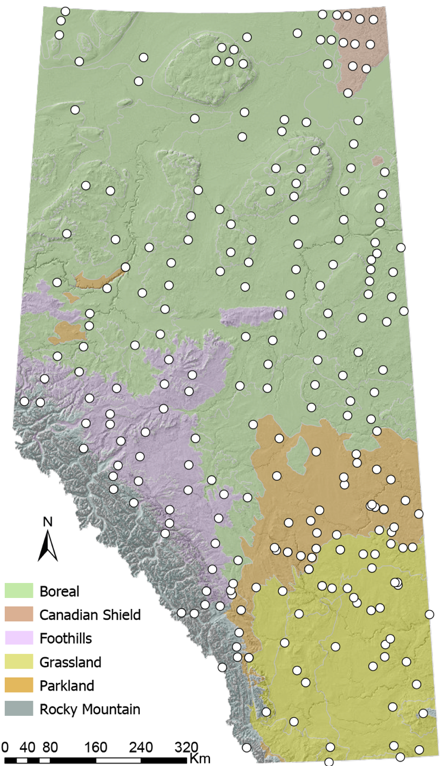
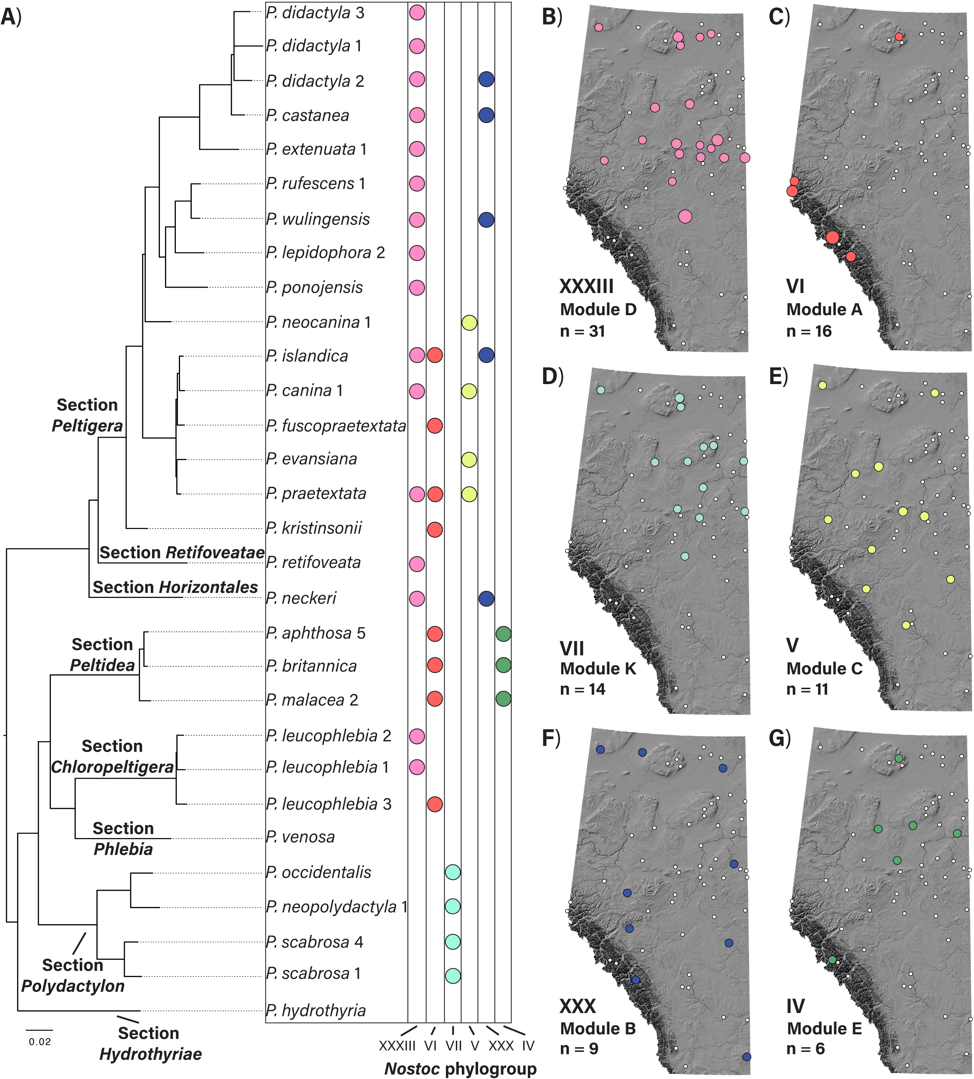
We are addressing this question by identifying the Peltigera species and their associated Nostoc phylogroups from a systematic sampling of over 3000 specimens collected by the Alberta Biodiversity Monitoring Institute (ABMI; Fig. 3). We are using this dataset to determine how the symbiotic interactions are structured by spatial gradients and across multiple scales (Fig. 4).
Figure 3. Sampling scheme across Alberta, Canada, with the six Natural Regions (biomes) of the province. Circles represent 246 sampling sites where the ABMI sampled over 3000 cyanolichen specimens, which are included in this study.
Figure 4. Association patterns and spatial distribution of the six most common Nostoc phylogroups associated with Peltigera in Alberta based on 157 thalli from 61 ABMI sampling sites. A) Maximum likelihood phylogeny of exemplar species representing the eight sections of the genus Peltigera, including 28 species found in association with the six most common Nostoc phylogroups in Alberta based on this preliminary sampling. Branch lengths are substitutions per site. Colored circles in the columns show the Nostoc phylogroups found in association with each species for this sampling. B-G) Spatial distribution of Nostoc phylogroups found in Peltigera thalli at 61 ABMI sampling sites (circles). Roman numerals correspond to Nostoc phylogroups. Colored circles represent sites where the six most common Nostoc phylogroups associated with Peltigera were found. The size of the colored circles denotes the number of thalli in which a Nostoc phylogroup was found at a given site. “n” is the total number of thalli in which a Nostoc phylogroup was found. Module labels correspond to the ones identified in Fig. 1.
Goal 1b. Does the availability of Nostoc phylogroups outside lichen thalli shape the local community of Nostoc in lichen thalli?
We will soon be conducting field work at multiple ABMI sites in Alberta to sample Peltigera cyanolichens along with their substrates in order to characterize the communities of lichenized and free-living Nostoc. This information will allow us to determine the role of Nostoc availability in shaping the communities of lichenized photobionts and mycobionts at fine spatial scales.
Goal 1c. Develop a tool to incorporate existing metrics of phylogenetic diversity into the assessment of symbiotic specificity and asymmetry in bipartite networks of interacting species.
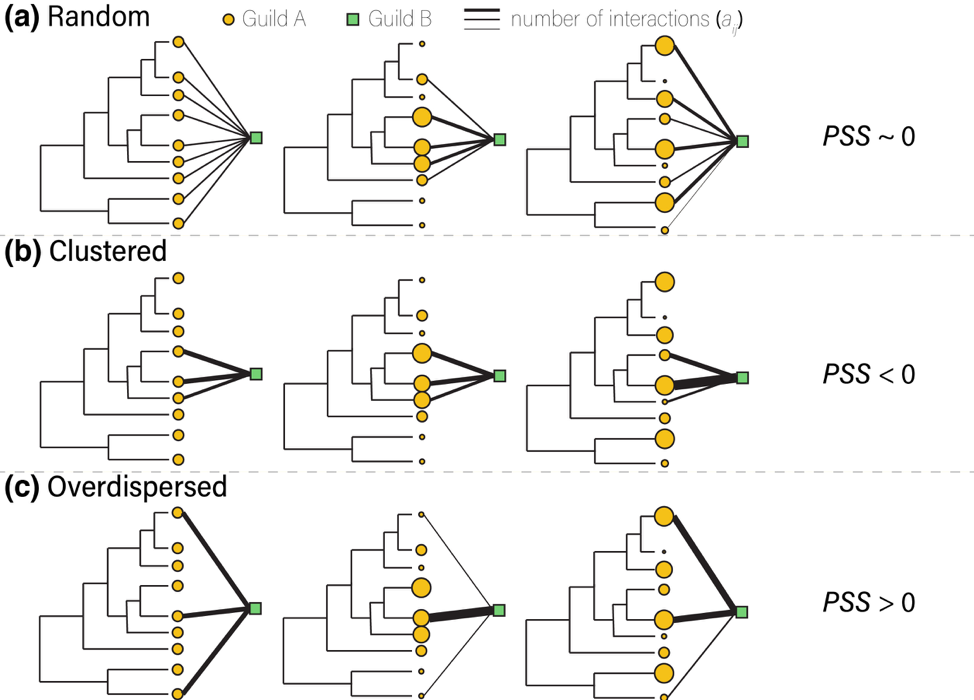
We developed a tool that integrates phylogenetic structure and partner availability to study specialization in interaction networks (Fig. 5). This tool will allow us to describe the ecological and evolutionary dimensions of specificity in the Peltigera–Nostoc association and their relationship with symbiont biodiversity patterns across multiple spatial scales.
Figure 5. Schematic representation of three possible patterns (a–c) of phylogenetic structure (random, clustered, and overdispersed) of partners (guild A) associated with one species from guild B. The size of the orange circles represents the relative availability of each member of guild A in an interaction matrix. Our approach measures the Phylogenetic Structure of Specialization (PSS) to characterize these different types of specialization. Adapted from Pardo-De la Hoz et al. (2020).
Goal 1d. Incorporate module composition uncertainty in modularity analyses.
We are developing a statistical tool to assess uncertainty in the estimation of module assignation in interaction network analyses.
Literature cited
Pardo-De la Hoz, C. J., Medeiros, I. D., Gibert, J. P., Chagnon, P.-L., Magain, N., Miadlikowska, J., Lutzoni, F. 2022. Phylogenetic structure of specialization: A new approach that integrates partner availability and phylogenetic diversity to quantify biotic specialization in ecological networks. Ecology and Evolution 12:e8649.