Most of the text for this section was written by the lead PI for the project, A. Elizabeth (Betsy) Arnold.
Our over-arching goal is to document the biodiversity of little-known endophytes associated with plants and lichens across broad temporal and spatial scales, with a focus on the vanishing biological landscape of the North American Arctic. The aims of this project are:
Aim 1: Survey preserved specimens of plants and lichens for endophytic fungi, in which herbarium specimens act as “snapshots” of symbiont communities. Using next-generation sequencing (NGS), we are able to use these “snapshots” to produce reliable data regarding endophytic and endolichenic diversity and composition of dried plants and lichens and their respective localities of origin for over the past 135 years (Fig. 1). We will sample ca. 350 plant and lichen herbarium specimens (Figs 2–3).

Fig. 1. Proof of concept: Endophyte communities in plant and lichen herbarium specimens represent a time capsule of endophyte diversity at the time of collection. (A) We examined endophytes in herbarium specimens of two plant species that occur along the boreal-to-arctic continuum: Andromeda polifolia and Ledum (Rhododendron) palustre. We extracted DNA from surface-sterilized tissue fragments and evaluated endophyte communities via a MiSeq-based workflow, with extensive positive and negative controls. In parallel, we cloned from subsets of the same tissue. Cloning provided similar signal to the MiSeq approach, but validated richness was ca. 8–12 times higher with the deeper sequencing by MiSeq. We obtained only one fungal isolate by culturing, underscoring the power of the culture-free approach. Richness via MiSeq was consistent with freshly collected specimens from similar environments. (B) We examined herbarium records and selected 300 focal arctic plant specimens with collection dates of 1884–2012. From each specimen, we need only a dime-sized piece of tissue (half of the black square), which can be gathered from tissue that has broken off from specimens over time (arrow points to fragment envelope). (C) In four-year-old specimens, diverse endophytic Pezizomycotina were observed via MiSeq: protruding bars between branches and the outer taxonomy ring indicate diverse endophytes from older material. Phylogenetic placement in T-BAS identified 24 orders and 38 families of fungi, consistent with fresh collections. (D) In parallel with Daru et al. (2018, ‡) our preliminary results (‡‡) are positive. Asterisks indicate discovery of diverse species and classes from herbarium specimens and that the most common classes of endophytes in fresh samples are represented in herbarium material preserved 100 and 118 years ago.
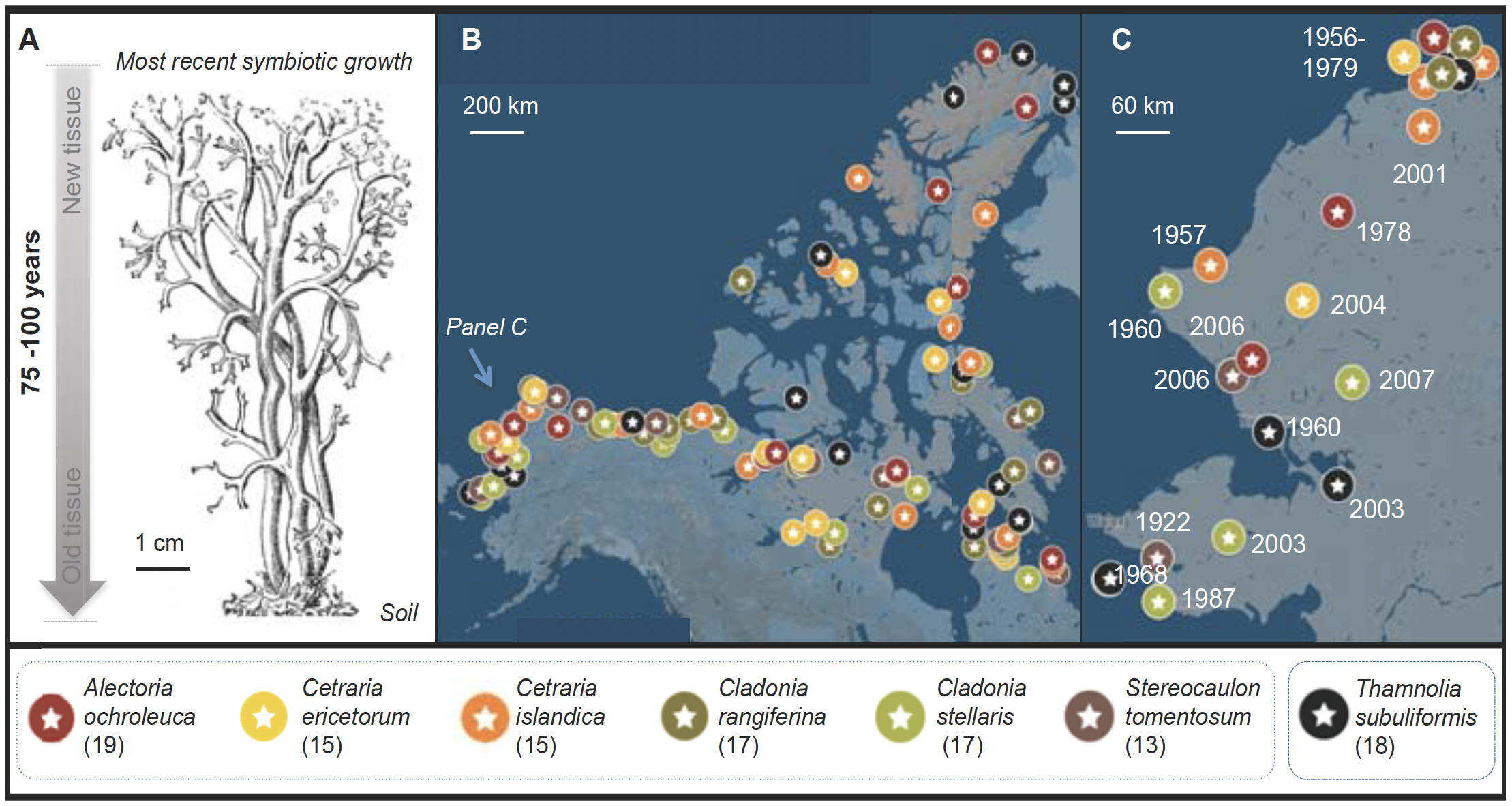
Fig. 2. (A) Fruticose lichen forms a chronosequence from the oldest tissue at the base to the newest at the tips. (B) We have approval for sampling ca. 150 lichen herbarium specimens for endophytes (subset shown here). (C) Detail, NW Alaska.
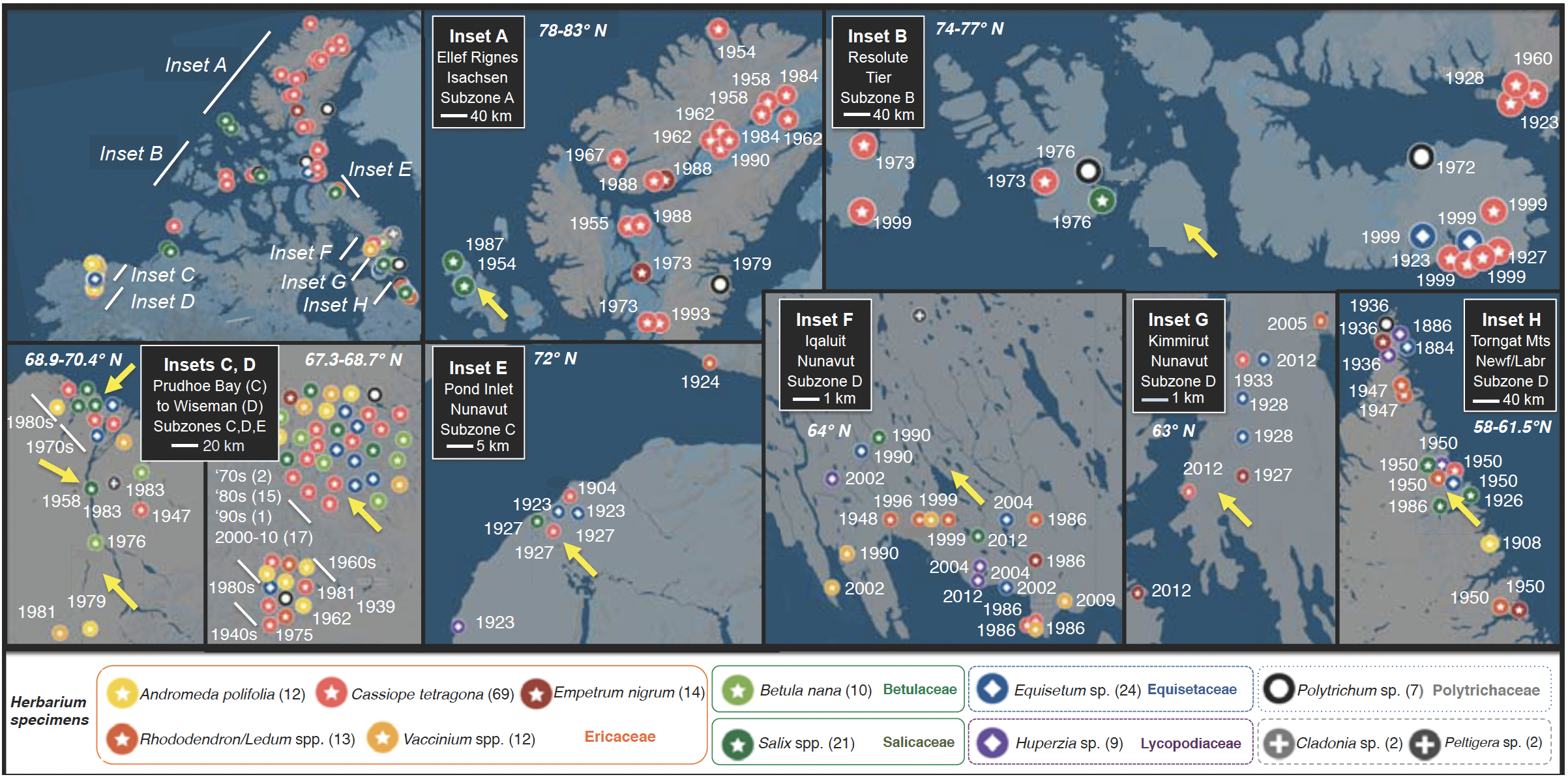
Fig. 3. Selection of plant herbarium specimens for examining endophyte communities, approved by partner herbaria. Map (upper left) shows Alaskan and Canadian Arctic, highlighting the geographic scope of sampling of the 185 representative plant specimens (of 300 selected for study). Regions with dense sampling are highlighted as A-H. Dates indicate year of collection Symbols correspond to host taxa (button panel). Numbers after taxonomic names indicate number of specimens selected per linage. Yellow arrows indicate locations for field campaigns (Aim 2) in which these and additional taxa will be sampled in Year 1 (Alaska, primary sites in insets C and D) and Year 2 (Canada, insets A, B, E–H) (cf. Fig. 4).
Aim 2: Survey endophytes systematically across transects of the major Arctic subzones in Alaska and Canada (Fig. 4). These surveys include new field collections in the same localities and host species complementary to the herbarium specimens of Aim 1 (Figs 2–3). We then use culture-free (NGS) and culture-based approaches to establish the biodiversity, host affiliations, composition, distributions, and β-diversity of endophyte communities. These data are compared to data from herbarium specimens in prediction-based frameworks.
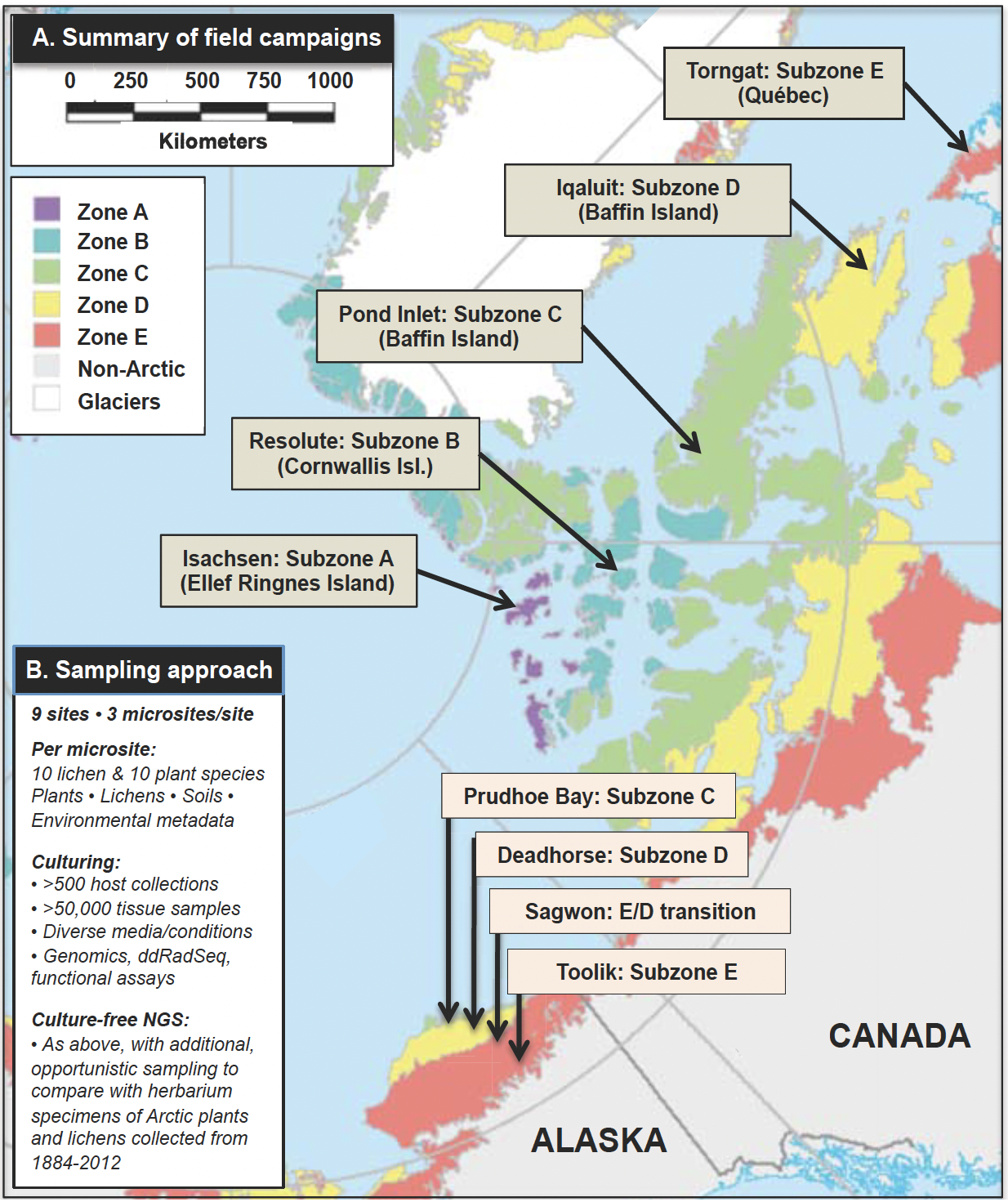
Fig. 4. (A) Summary of the field campaigns: Alaska (Year 1) and Canada (Year 2), with details of the scope of the sampling effort in (B).
Aim 3: Extend and share tools for biodiversity informatics to accelerate discovery and systematics for fungal biodiversity from culture-based and NGS studies (Fig. 5). Specifically, we aim to deliver robust multilocus reference trees (enriched with previously unknown endophytes) for the hyperdiverse phylum Ascomycota. These trees are able to detect shifts in host specificity, rates of diversification, and ecological modes. Finally, we will integrate population, genome, ecological, and phylogenetic data to describe new taxa, focusing on hyper-diverse clades Coniochaeta and Daldinia.
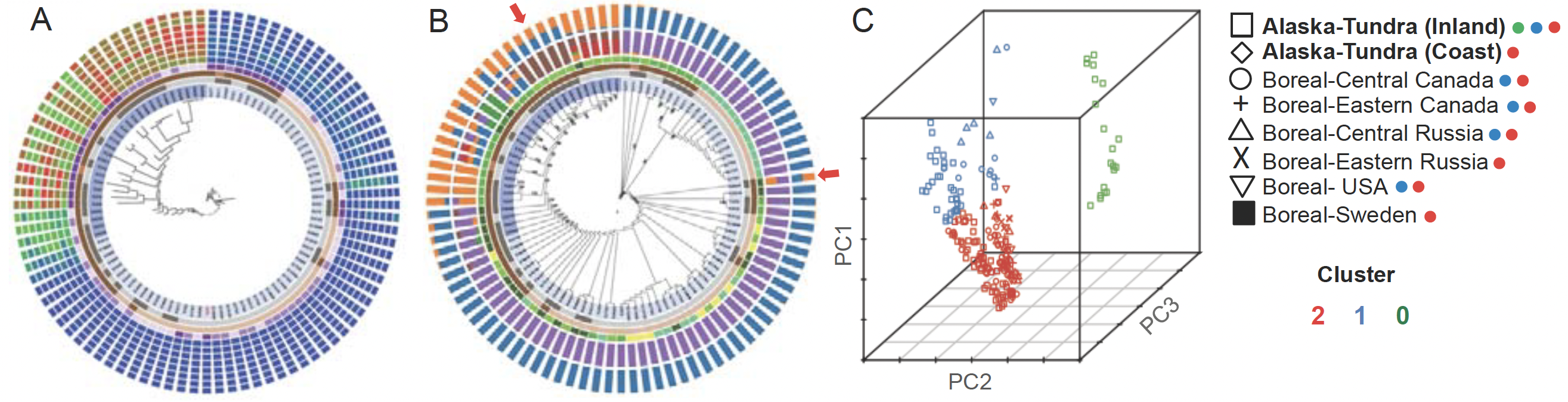
Fig. 5. Advancing T-BAS to integrate ddRADseq and low coverage genome data will illuminate phylogenetic incongruence in signal from different chromosomes, supercontigs, or loci, and the degree of admixture among individuals and populations. (A) Prototype display for a model clonal pan-genome (two lineages are obvious). In the smaller lineage is a recombining pan-genome (different genetic backgrounds are discernable). (B) Evidence of genetic admixture between the lineages shown in (A): two outermost rings quantify admixture via two methods (LnP(D) and delta K). Histogram bars in the outer ring (from delta K) show that some individuals (arrows) are recently admixed with both lineages (blue and orange) represented in their ancestry. (C) Principal component analysis of 292 Daldinia samples via ddRADseq, showing multiple evolutionary forces shaping contemporary populations: (1) at least three distinct genetic clusters; (2) local structuring in Alaska tundra (blue and green); (3) long distance dispersal in cluster 2; and (4) evidence of restricted gene flow in cluster 0 inferred from the large vertical dispersal of the inland Alaskan sample. Further examination of ddRADseq data for the presence of MAT1-1/MAT1-2 mating type idiomorphs in the Alaskan Daldinia populations shows a strong skew to MAT1-2, containing a MATA_HMG domain (high-mobility group), suggesting a predominantly asexual mode of reproduction.
Literature Cited
Daru, B.H., Bowman, E.A., Pfister, D., and Arnold, A.E. 2018. A novel proof-of-concept for capturing the diversity of endophytic fungi preserved in herbarium specimens. Philosophical Transactions of the Royal Society B 374: 20170395.