Symbiotic interactions, including parasitism, commensalism, and mutualism, form complex networks that are ubiquitous in nature. This study aims to understand the processes that shape those networks using cyanolichens, which are iconic symbioses consisting mainly of mutually beneficial fungi and cyanobacteria. To reach this goal, we are implementing a fully integrated approach combining evolutionary biology and ecology.
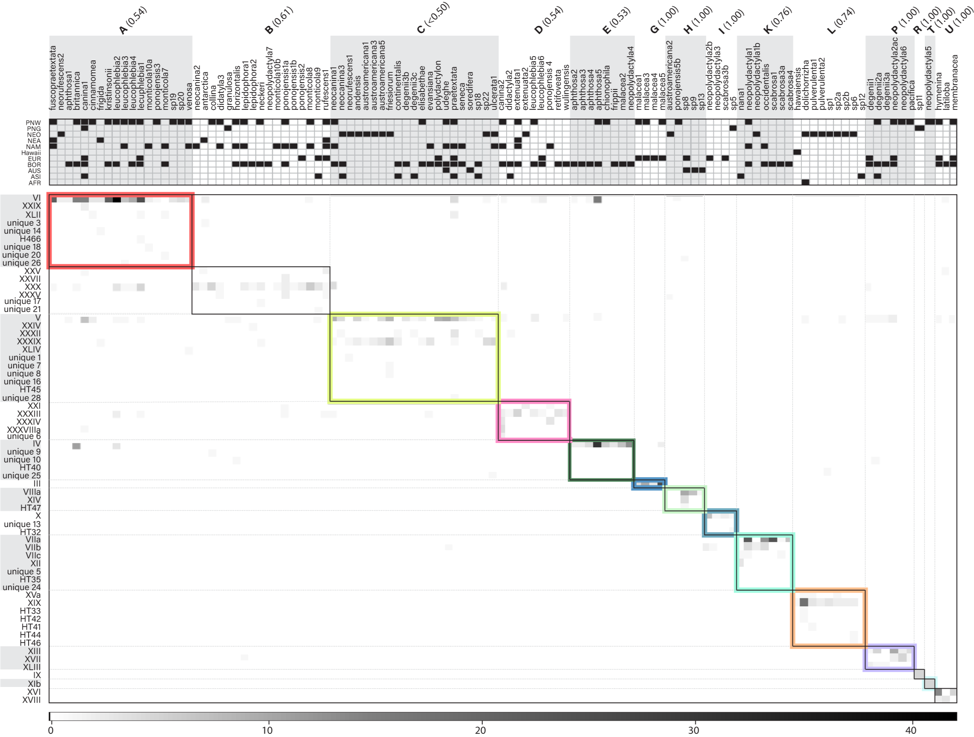
Figure 1. Interaction matrix and modules uncovered through a global sampling of Peltigera. In the main matrix, shades of grey are proportional to the number of thalli in which a given Peltigera species (columns) was found in association with a corresponding Nostoc phylogroup (rows). The black boxes (often with color highlights) delineate the modules found through simulated annealing. Grey dotted lines and alternating white and grey backgrounds outside the matrix were used to facilitate visual allocation of given cyanobionts and mycobionts to a given module. On the top panel, we show the presence/absence of Peltigera species in each of the 11 sampled biogeographical regions (AFR = Africa, ASI = Asia, AUS = Australasia, BOR = Boreal biome, EUR = Europe, NAM = North America, NEA = Argentina and Chile, NEO = Neotropics, PNG = Papua New Guinea, PNW = Pacific Northwest). The same was done for Nostoc phylogroups but was removed because of lack of space. Module IDs (letters in bold, top panel) are shown with module frequency/support values shown in parentheses. Module IDs and colors correspond to those shown in Fig. 2. Adapted from Chagnon et al. (2019).
Recent studies of the lichen model genus Peltigera and its Nostoc symbionts have revealed: (i) that Peltigera species are mostly specialized on generalist Nostoc phylogroups (Fig. 1), (ii) that the interactions and the high level of specialization by the fungus are phylogenetically conserved (Fig. 2), and (iii) that bioclimatic factors, at an intra-biome (boreal) scale, are more limiting than cyanobiont availability for explaining geographic distributions of Peltigera species.
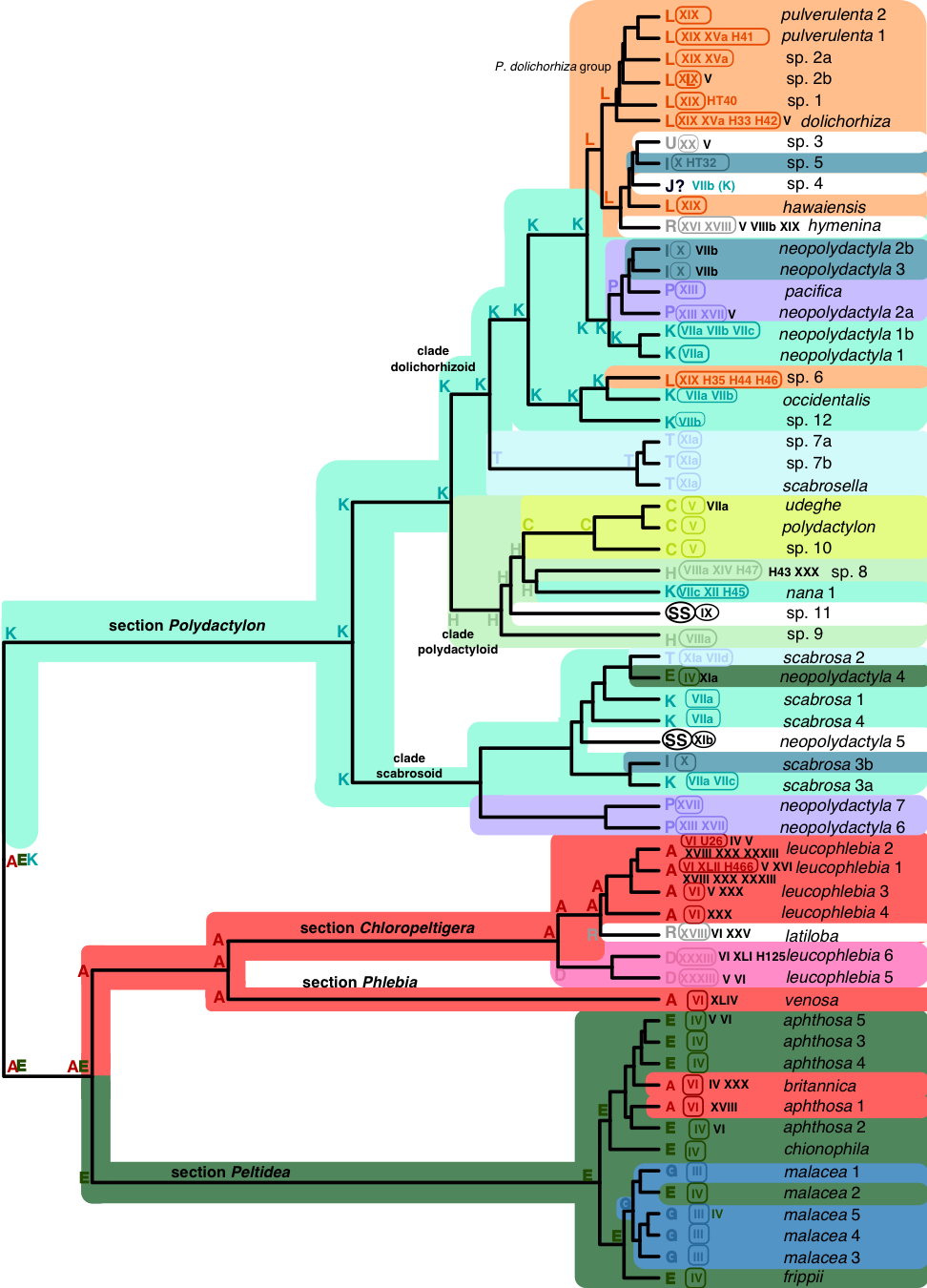
Figure 2. Seven-locus tree (relative time) of four sections of the genus Peltigera. Each terminal branch represents a putative species listed on the far right. The colored capital letters immediately to the right of the terminal branches represent network modules, based on results of the modularity analysis (Fig. 1). Further to the right, the Nostoc phylogroup partners of each Peltigera species are shown. Each Nostoc phylogroup belonging to the same module as its Peltigera species partner, shares the same color inside a rounded box. Nostoc phylogroups belonging to different modules are shown in black outside of the rounded boxes. Inferences of ancestral modules on internal nodes were generated using BioGeoBears with the DEC+J model (Matzke 2013). Background colors represent the current and inferred ancestral modules for each Peltigera species and ancestral lineages. The same colors were used in Fig. 1. Small, low frequency modules have no background colors. Strict 1:1 specialist associations forming distinct modules are represented in a circle with “SS”. Modules represented with a question mark are presumably incorrectly reconstructed by the modularity analyses (species in modules with none of their partners) and the module of their cyanobiont was used for the color-coding scheme. Branches and nodes with very low probabilities for all or many modules have no background color. Adapted from Chagnon et al. (2019).
This study will capitalize on the large-scale, systematic, and multi-taxon monitoring program of the Alberta Biodiversity Monitoring Institute (ABMI). This project represents a unique opportunity to test three important drivers of symbiotic network assembly:
(1) Spatial scales and gradients: Are interactions well predicted by environmental biotic and abiotic properties, and/or spatial overlap due to stochastic dispersal?
(2) Phylogenetic history: Do Nostoc symbionts show phylogenetic conservatism in their selection of Peltigera partners, as shown by Peltigera towards their Nostoc partners?
(3) Specific recognition molecules: Are Nostoc genes differentially expressed in lichen thalli vs. a free-living state, and does their genotypic variation explain the observed specificity between symbiotic partners in nature?
Literature cited
Chagnon, P. L., Magain, N., Miadlikowska, J., & Lutzoni, F. (2019). Species diversification and phylogenetically constrained symbiont switching generated high modularity in the lichen genus Peltigera. Journal of Ecology, 107(4), 1645-1661.
Matzke, N.J. (2013). Probabilistic historical biogeography: New models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing. Frontiers of Biogeography, 5, 242-248.