The global phylogeny will be based exclusively on sequence data from nuclear ribosomal ITS, LSU, nuclear RPB1 and beta-tubulin, which will allow the inference of the evolution of phenotypic traits.
The goal of this phenotypic study will be to find synapomorphies for Peltigera sections and species, including photobiont switches. Because the resulting phylogenetic tree will include up to 350 specimens representing the geographical and phenotypical diversity of all known and new putative species of Peltigera, we will be able to determine if part of the Peltigera phylogenetic structure contains a geographical component. If so, a more in-depth historical biogeography analysis of this genus will be performed.
The same historical biogeographical analysis will be possible on the rbcLXS phylogeny of the Nostoc found in these 350 lichens.
Our reconstruction of the phylogeny of both symbionts for up to 350 specimens will also enable us to study the interplay between symbiotic selectivity and geography during the evolution of Peltigera.
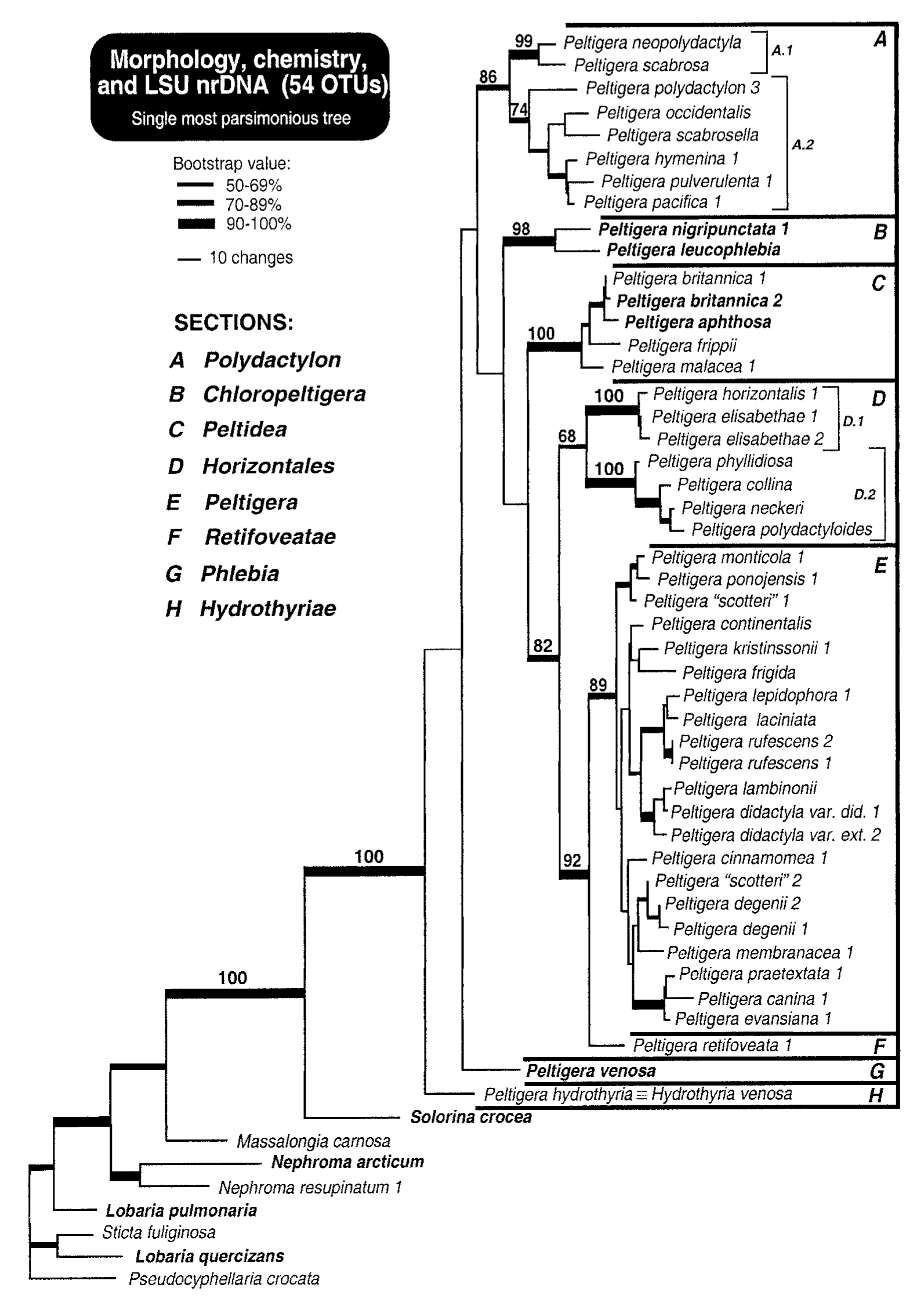 |
Fig. 2. Single most parsimonious tree resulting from a combined analysis of chemical, morphological, and LSU nrDNA data for a subsample of 54 OTUs. Tree length 1429.0 steps, CI (excluding uninformative characters) p 0.682, RIp 0.819. Bootstrap values greater than 50% are shown with increasingly thicker branches. Bootstrap values supporting the main monophyletic entities revealed by this study are given above these critical internodes. Groups A–H correspond to the same infrageneric sections as shown in figs. 6 and 7. A.1, A.2 and D.1, D.2 represent main clades within sections A and D, respectively. Names in bold indicate trimembered symbiotic entities. (Miadlikowska and Lutzoni, 2000)