Our main objective is to infer if Nostoc lineages show phylogenetic conservatism in the selection of Peltigera species. We will achieve this through two specific goals:
Goal 2a. Infer a phylogeny for Nostoc Clade II using multiple loci and accounting for horizontal gene transfers.
Confidently inferring the evolutionary relationships among cyanobacteria is challenging because of the prevalence of Horizontal Gene Transfer (HGT). We have identified loci that we hypothesize to be less vulnerable to HGT and improve phylogenetic support across Nostocales (Fig. 6). We are currently sequencing dozens of cyanolichens metagenomes and extracting these loci to generate a phylogenomic framework for Nostoc Clade II, which includes most lineages of lichenized cyanobacteria.
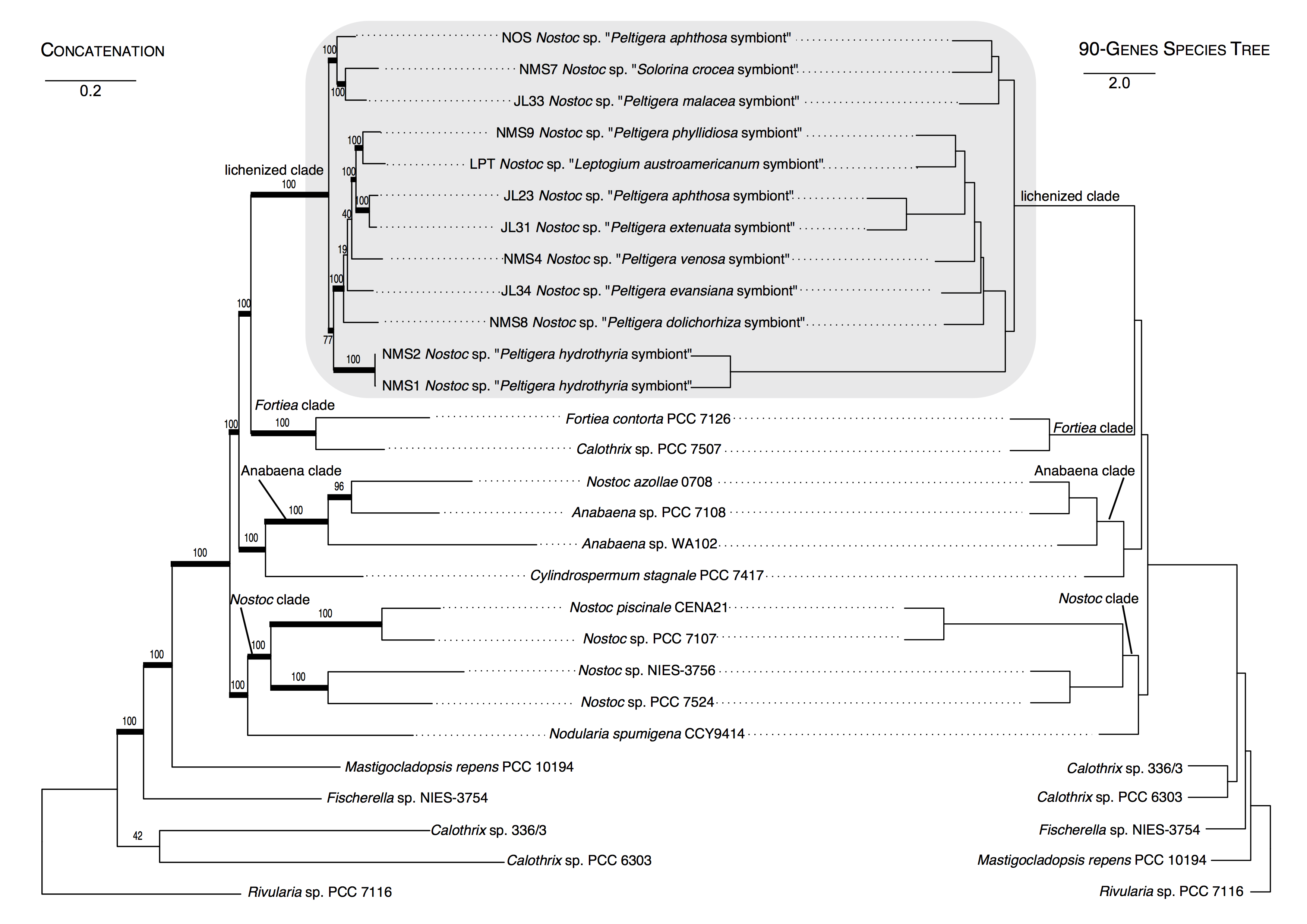
Figure 6. Comparison of the topologies for Nostocales resulting from a ML analysis using a concatenated 90-locus dataset (left) and from a 90-gene species tree approach (right). The outgroup is Rivularia sp. (PCC 7116). Thick branches on the concatenated tree represent bootstrap support ≥70%. Scales = nucleotide substitutions per site for the concatenated tree, and coalescence units for the species tree. The grey shaded box highlights the clade of cyanobacteria found in associations with species of Peltigerales.
Goal 2b. Infer the evolution of network modules on the Nostoc phylogeny to test for phylogenetic conservatism.
We will use the phylogenetic framework from goal 2a to determine whether closely related lineages of Nostoc are part of the same modules within the interaction network.