Liu et al. (1999) designed a number of fungal-specific primers from this molecule. Later, Reeb et al. (2004) developed 11 more primers in order to resolve phylogenetic relationships between lichenized fungi. Recently, members of our lab developed two more primers that are specific to lichenized fungi in the Peltigerales (Hofstetter et al. 2007). Please let us know if new primers that we are not aware of should be included here.
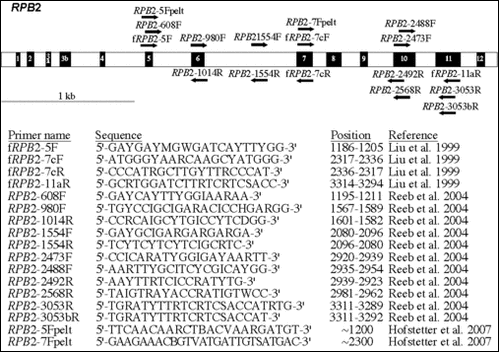 |
The above image is a representation of the RPB2 gene according to Liu et al. (1999). The black numbered boxes represent the 12 amino acid regions that are conserved throughout eukaryotes. Positions of the RPB2 primers used for symmetric PCR amplifications and cycle sequencing reactions are represented by black arrows. Primer names in the table are followed by their degenerate oligonucleotide sequences, their positions relative to the RPB2 gene sequence of Saccharomyces cerevisiae (GenBank Accession No. M15693), and their respective sources.
Kraichak et al. (2015) developed two RPB2 primers designed specifically for Graphidaceae.
GD1-RPB2-7cF (5′- GAG CGA ATG GAT ACC ATG GCG A -3′)
GD-RPB2-11aR (5′- GCT TAC GCC CGG TGT GAC CAT TGT -3′)
Medeiros et al. (2021) developed new RPB2 primers for Lecanoraceae. The first of these, lecRPPB2-6F, was designed to be paired with fRPB2-7cR in the second round of a nested PCR, after fRPB2-5F and fRPB2-7cR were used for the first amplification. The second new primer, lecRPB2-seq7R, was designed as a Sanger sequencing primer to replace fRPB2-7cR.
lecRPPB2-6F 5′- TGG GGN YTR GTM TGY CCD GC -3′
lecRPB2-seq7R 5′- G RTT RTG RTC NGG RAA NGG -3′
References:
Kraichak, E. Lücking, R., Aptroot, A., Beck, A., Dornes, P., John, V., Lendemer, J.C., Nelsen, M.P., Neuwirth, G., Nutakki, A., Parnmen, S., Sohrabi, M., Tønsberg, T. and Lumbsch, H.T. 2015. Hidden diversity in the morphologically variable script lichen (Graphis scripta) complex (Ascomycota, Ostropales, Graphidaceae). Organisms Diversity & Evolution 15(3): 447-458.
Liu, Y.J., Whelen, S. and Hall, B.D. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16(12): 1799-1808.
Medeiros, I.D., Mazur, E., Miadlikowska, J., Flakus, A., Rodriguez-Flakus, P., Pardo-De la Hoz, C.J., Cieślak, E., Śliwa, L., and Lutzoni, F. 2021. Turnover of lecanoroid mycobionts and their Trebouxia photobionts along an elevation gradient in Bolivia highlights the role of environment in structuring the lichen symbiosis. Frontiers in Microbiology 12: 774839.
Download publication (PDF file)
Reeb, V., Lutzoni, F. and Roux, C. 2004. Contribution of RPB2 to multilocus phylogenetic studies of the Pezizomycotina (euascomycetes, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Molecular Phylogenetics and Evolution 32:1036-1060.
Download publication (PDF file)
Hofstetter, V., Miadlikowska, J., Kauff, F., and Lutzoni, F. 2007. Phylogenetic comparison of protein-coding versus ribosomal RNA-coding sequence data: A case study of the Lecanoromycetes (Ascomycota). Molecular Phylogenetics and Evolution 44:412-426.
Download publication (PDF file)