We used the primer list from Vilgalys’ lab at Duke University as the basis to which we have added primers developed in our lab. Contrary to Vilgalys’ lab who are designing primers mainly for phylogenetic studies of Basidiomycota, the target organisms for our primers are typically lichenized and non-lichenized Ascomycota. Primers that work best for us are highlighted in blue. Many of these primers are likely to work more generally (e.g., for basal fungal lineages). Several of these primers work well on plants, as well as on algae found in lichens. Recently, members of our lab developed species-specific primers for nuclear ribosomal loci of Verrucariaceae (Gueidan et al. 2007).
Link to: Vilgalys’ lab at Duke University
– Click on image to enlarge –
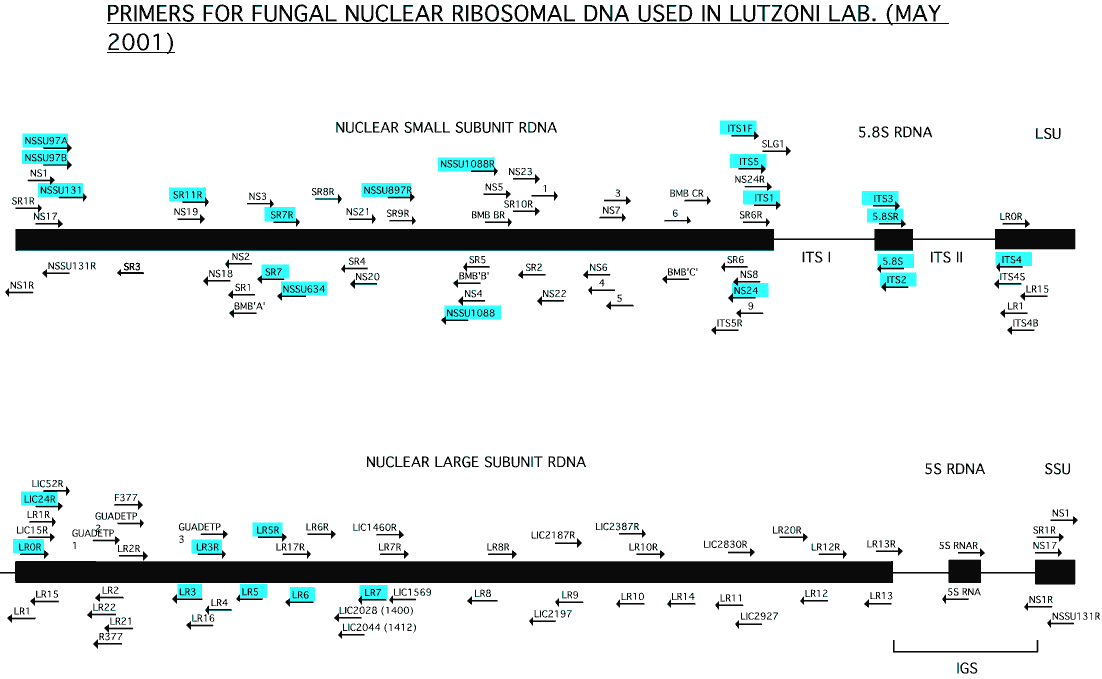 |
| Small subunit rDNA primer sequences | ssu ref. | nb pb | |
| BMB-‘A’ (Lan) | GWA TTA CCG CGG CKG CTG | 583-566 | 18 |
| BMB-‘B’ (Lan) | CCG TCA ATT CMT TTR AGT TT | 1147-1128 | 20 |
| BMB-‘C’ (Lan) | ACG GGC GGT GTG TRC | 1641-1637 | 15 |
| BMB-BR (Lan) | CTT AAA GGA ATT GAC GGA A | 1131-1149 | 19 |
| BMB-CR (Lan) | GTA CAC ACC GCC CGT CG | 1637-1643 | 17 |
| nssu97a (Kau) | TAT ACG GTG AAA CTG CGA ATG GC | 75-97 | 23 |
| nssu97b (Kau) | CGG TGA AAC TGC GAA TGG C | 79-97 | 19 |
| nssu634 (Kau) | CCC CAG AAG GAA AGI CCC GIC C | 705-684 | 22 |
| nssu897R (Kau) | AGA GGT GAA ATT CTT GGA | 897-914 | 18 |
| nssu1088 (Kau) | TGA TTT CTC GTA AGG TGC CG | 1088-1069 | 20 |
| nssu1088R (Kau) | CGG CAC CTT ACG AGA AAT CA | 1069-1088 | 20 |
| nssu131 (Kau) | CAG TTA TCG TTT ATT TGA TAG TAC C | 107-131 | 25 |
| NS1 (Whi) | GTA GTC ATA TGC TTG TCT C | 20-38 | 19 |
| NS2 (Whi) | GGC TGC TGG CAC CAG ACT TGC | 573-553 | 21 |
| NS3 (Whi) | GCA AGT CTG GTG CCA GCA GCC | 553-573 | 21 |
| NS4 (Whi) | CTT CCG TCA ATT CCT TTA AG (sim. to BMB-b) | 1150-1131 | 20 |
| NS5 (Whi) | AAC TTA AAG GAA TTG ACG GAA G (sim. to BMB-BR) | 1129-1150 | 22 |
| NS6 (Whi) | GCA TCA CAG ACC TGT TAT TGC CTC | 1436-1413 | 24 |
| NS7 (Whi) | GAG GCA ATA ACA GGT CTG TGA TGC | 1413-1436 | 24 |
| NS8 (Whi) | TCC GCA GGT TCA CCT ACG GA | 1788-1769 | 20 |
| NS17 (Gar) | CAT GTC TAA GTT TAA GCA A | 54-72 | 19 |
| NS18 (Gar) | CTC ATT CCA ATT ACA AGA CC | 516-497 | 20 |
| NS19 (Gar) | CCG GAG AAG GAG CCT GAG AAA C | 381-402 | 22 |
| NS20 (Gar) | CGT CCC TAT TAA TCA TTA CG | 871-852 | 20 |
| NS21 (Gar) | GAA TAA TAG AAT AGG ACG | 802-819 | 18 |
| NS22 (Gar) | AAT TAA GCA GAC AAA TCA CT | 1312-1297 | 20 |
| NS23 (Gar) | GAC TCA ACA CGG GGA AAC TC | 1184-1203 | 20 |
| NS24 (Gar) | AAA CCT TGT TAC GAC TTT TA | 1769-1750 | 20 |
| SR1 (VilU) | ATT ACC GCG GCT GCT (similar to BMB-‘A’) | 581-567 | 15 |
| SR1R (Vil0) | TAC CTG GTT GAT TCT GC | 21-1 | 21 |
| SR2 (Elw) | CGG CCA TGC ACC ACC | 1277-1263 | 15 |
| SR3 (VilU) | GAA AGT TGA TAG GGC T | 321-306 | 16 |
| SR4 (VilU) | AAA CCA ACA AAA TAG AA | 841-825 | 17 |
| SR5 (VilU) | GTG CCC TTC CGT CAA TT | 1155-1139 | 17 |
| SR6 (VilU) | TGT TAC GAC TTT TAC TT | 1763-1747 | 17 |
| SR7 (VilU) | GTT CAA CTA CGA GCT TTT TAA | 637-617 | 21 |
| SR7R (VilU) | TTA AAA AGC TCG TAG TTG AAC | 617-637 | 21 |
| SR8R (VilU) | GAA CCA GGA CTT TTA CCT T | 732-750 | 19 |
| SR9R (Elw) | YAG AGG TGA AAT TCT | 896-910 | 15 |
| SR10R (VilU) | TTT GAC TCA ACA CGG G | 1181-1196 | 16 |
| SR11R (Spa) | GGA GCC TGA GAA ACG GCT AC | 389-408 | 20 |
| 1 (DeP) | GCT CTT TCT TGA TTT TGT | 1241-1258 | 18 |
| 3 (DeP) | TTT GAG GCA ATA ACA GGT | 1410-1427 | 18 |
| 4 (DeP) | CAT CTA AGG GCA TCA CAG | 1445-1428 | 18 |
| 5 (DeP) | TTG TCT CTG TCA GTG TAG | 1482-1465 | 18 |
| 6 (DeP) | CAA CGA GGA ATT CCT AGT | 1566-1583 | 18 |
| 9 (DeP) | AAT GAT CCT TCC GCA GGT | 1797-1780 | 18 |
| Large subunit rDN primer sequences | lsu ref. | nb pb | |
| GUADET P1 (Gua) | GAG TCG AGT TGT TTG GGA ATG CAG C | 275-300 | 25 |
| GUADET P2 (Gua) | GAA AAG AAC TTT GAA AAG AGA GTG AA | 379-403 | 26 |
| GUADET P3 (Gua) | CCC GTC TTG AAA CAC GGA CCA AGG | 661-685 | 24 |
| P2R (Reh) | CTC TCT TTT CAA AGT TCT TTT CAT CT | 1412-1396 | 17 |
| LR0R (VilU) | ACC CGC TGA ACT TAA GC | 26-42 | 17 |
| LR0R (Reh) | GTA CCC GCT GAA CTT AAG C | 24-42 | 19 |
| LR1R (Mon) | AGG AAA AGA AAC CAA CC | 57-73 | 17 |
| LR2 | TTT TCA AAG TTC TTT TC | 395-379 | 17 |
| LR2R | AAG AAC TTT GAA AAG AG | 382-398 | 17 |
| LR3 (Vil0) | GGT CCG TGT TTC AAG AC | 677-661 | 17 |
| LR3R | GTC TTG AAA CAC GGA CC | 664-680 | 17 |
| LR4 | ACC AGA GTT TCC TCT GG | 887-871 | 17 |
| LR5 (Vil0) | ATC CTG AGG GAA ACT TC | 997-981 | 17 |
| LR5R (VilU) | GAA GTT TCC CTC AGG AT | 981-997 | 17 |
| LR6 (Vil0) | CGC CAG TTC TGC TTA CC | 1173-1157 | 17 |
| LR7 (Vil0) | TAC TAC CAC CAA GAT CT | 1483-1467 | 17 |
| LR7R (Vil0) | AGA TCT TGG TGG TAG TA | 1465-1481 | 17 |
| LR8 | CAC CTT GGA GAC CTG CT | 1917-1901 | 17 |
| LR8R | AGC AGG TCT CCA AGG TG | 1901-1917 | 17 |
| LR9 | AGA GCA CTG GGC AGA AA | 2292-2276 | 17 |
| LR10 | AGT CAA GCT CAA CAG GG | 2508-2492 | 17 |
| LR10R | GAC CCT GTT GAG CTT GA | 2490-2506 | 17 |
| LR11 | GCC AGT TAT CCC TGT GGT AA | 2919-2900 | 20 |
| LR12 (Vil0) | GAC TTA GAG GCG TTC AG | 3221-3204 | 17 |
| LR12R (VilU) | CTG AAC GCC TCT AAG TCA GAA | 3205-3225 | 21 |
| LR13 | CAT CGG AAC AAC AAT GC | 3464-3448 | 17 |
| LR14 | AGC CAA ACT CCC CAC CTG | 2701-2694 | 18 |
| LR15 | TAA ATT ACA ACT CGG AC | 161-145 | 17 |
| LR16 (Mon) | TTC CAC CCA AAC ACT CG | 716-700 | 17 |
| LR17R | TAA CCT ATT CTC AAA CTT | 1066-1083 | 18 |
| LR20R | GTG AGA CAG GTT AGT TTT ACC CT | 3058-3080 | 23 |
| LR21 | ACT TCA AGC GTT TCC CTT T | 442-424 | 19 |
| LR22 | CCT CAC GGT ACT TGT TCG CT | 371-352 | 20 |
| F377 | AGA TGA AAA GAA CTT TGA AAA GAG AA | 375-400 | 26 |
| R377 | CTC TCT TTT CAA AGT TCT TTT CAT CT | 400-375 | 26 |
| LIC15R (Mia2) | GGA GGA AAA GAA ACC AAC AG | 55-74 | 20 |
| * LIC24R (Mia1) | GAA ACC AAC AGG GAT TG | 64-80 | 17 |
| LIC2044 (Kau) | ACG CCT GCC TAC TCG CC | 26-42 | 17 |
| IGS and 5S rDNA p rimer sequences | |||
| 5S RNA (VilU) | ATC AGA CGG GAT GCG GT | 17 | |
| 5S RNAR (VilU) | ACQ GCA TCC CGT CTG AT | 17 | |
| NS1R | GAG ACA AGC ATA TGA CTA C | 38-20 (SSU) | 19 |
| LR13R | GCA TTG TTG TTC CGA TG | 3448-3464(lsu) | 17 |
| ITS and 5.8S rDNA primer sequences | |||
| ITS1 (Whi) | TCC GTA GGT GAA CCT GCG G | 1769-1787(ssu) | 19 |
| ITS1F (Gad) | CTT GGT CAT TTA GAG GAA GTA A | 22 | |
| ITS2 (Whi) | GCT GCG TTC TTC ATCG ATG C (sim. to 5.8S) | 20 | |
| ITS3 (Whi) | GCA TCG ATG AAG AAC GCA GC (sim. to 5.8SR) | 20 | |
| ITS4 (Whi) | TCC TCC GCT TAT TGA TAT GC | 60-41(lsu) | 20 |
| ITS4B (Gad) | CAG GAG ACT TGT ACA CGG TCC AG | 23 | |
| ITS4S (Kre) | CCT CCG CTT ATT GAT ATG CTT AAG | 59-36 (lsu) | 24 |
| ITS5 (Whi) | GGA AGT AAA AGT CGT AAC AAG G (sim. to SR6R) | 1745-1766(ssu) | 22 |
| ITS5R | CCT TGT TAC GAC TTT TAC TTC C | 1766-1745 (ssu) | 22 |
| 5.8S (Vil0) | CGC TGC GTT CTT CAT CG | 17 | |
| 5.8SR (Vil0) | TCG ATG AAG AAC GCA GC | 18 | |
| SR6R (VilU) | AAG TAP AAG TCG TAA CAA GG | 1747-1766(ssu) | 20 |
| LR1 (Vil0) | GGT TGG TTT CTT TTC CT | 73-57(lsu) | 17 |
| SLG-1 (Gro) | TTG CGC AAC CTG CGG AAG GAT | 1773-1793(ssu) | 21 |
| NS24R | TAA AAG TCG TAA CAA GGT TT | 1750-1769(ssu) | 20 |
| ssu ref. Mankin et al., 1986. Gene 44:143-145 lsu ref. Lapeyre et al., 1993. Nucleic Acids Res. 21:3322-3322 |
| * Primers designed in Lutzoni’s lab (DeP) DePriest,1993. Gene 134:67-74 (Elw) Elwood et al., 1985. Mol. Biol. Evol. 2:399-410 (Gad) Gardes and Bruns, 1993. Mol. Ecol. 2:113-118 (Gar) Gargas and Taylor, 1992. Mycologia 84(4): 589-592 (Gro) Groner and LaGreca, 1997. Lichenologist 29(5):441-454 (Gua) Guadet et al., 1989. Mol. Biol. Evol. 6:227-242 (Kau) Kauff and Lutzoni, 2002. Mol. Phylogen. Evol. 25:138-156 (Kre) Kretzer et al., 1996. Mycologia 88(5): 776-785 (Lan) Lane et al., 1985. PNAS USA 82:6955-6959 (Mia1) Miadlikowska and Lutzoni, 2000. Int. J. Plant Sci. 161(6) (Mia2) Miadlikowska, McCune, Lutzoni,2001. Bryologist 105:1-10 (Mon) Moncalvo et al., 1993. Mycologia 85(5):788-794 (Reh) Rehner and Samuels, 1994. Mycol. Res. 98(6):625-634 (Spa) Spatafora et al, 1995. J. Clinical Microbiol. 33:1322-1326 (Vil0) Vilgalys and Hester, 1990. Journal of bacteriology 172:4238-4246 (VilU) Vilgalys unpublished [http://www.botany.duke.edu/fungi/mycolab] (Whi) White et al., 1990. pp 315-322 in PCR protocols: A guide to methods and applications |
New Primers for Verrucariaceae
Species-specific primers for nuclear ribosomal loci of Verrucariaceae (Gueidan et al. 2007):
Download list (PDF file)